AgnostiMap: Proteome-Wide Interaction Discovery
Eliminate off-target blindspots by mapping where your molecule sequesters in the human proteome
About the Project
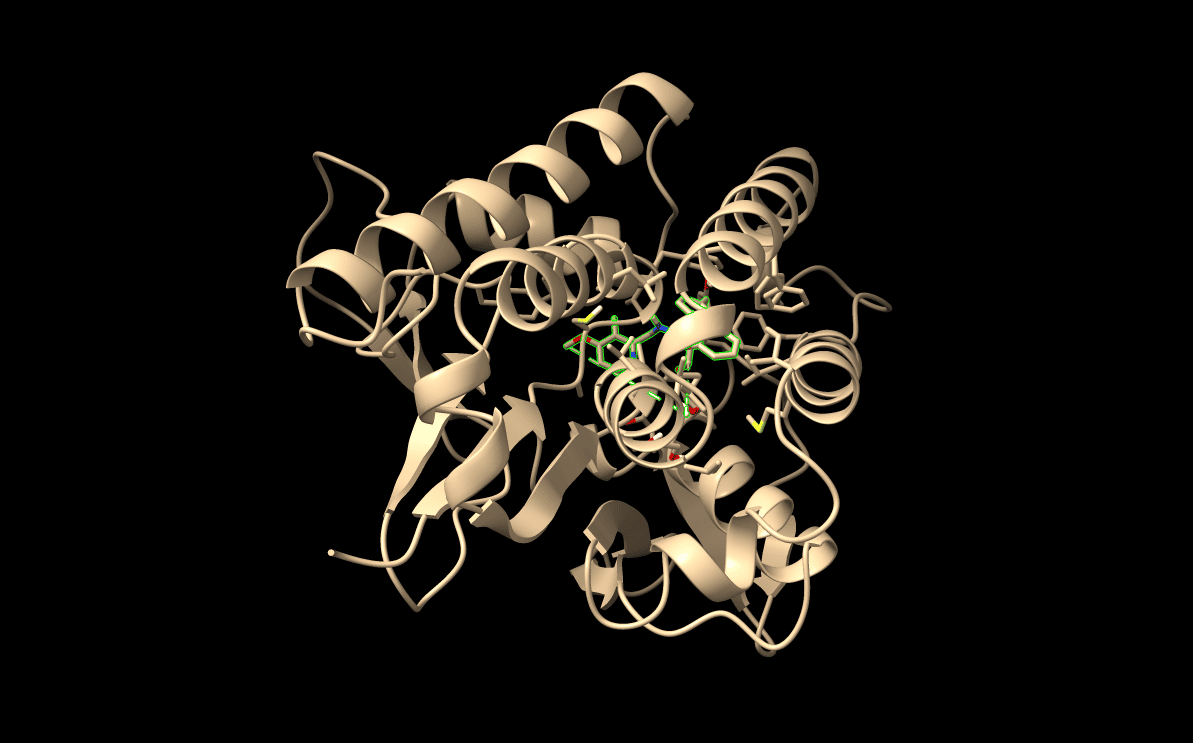
AgnostiMap is a target-agnostic drug–proteome mapping platform designed to reveal how small molecules interact with the full human proteome. Using AlphaFold 3 (v6) derived structures, the system screens candidate ligands against more than 20,000 proteins without prior target assumptions.
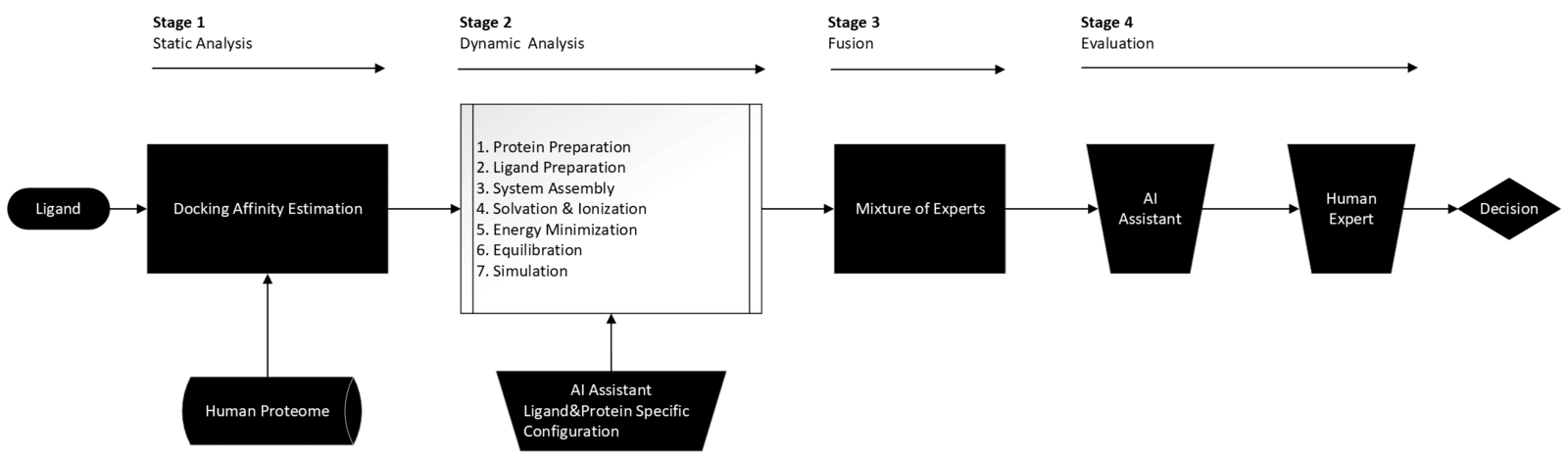
AgnostiMap operates through a rigorous, multi-stage pipeline:
High-Throughput Screening: Systematic docking estimations across the entire proteome to identify potential binding sites.
MD Validation:Molecular dynamics simulations (10 ns+) to test binding persistence and cavity stability under realistic motion.
AI-Driven Fusion: Fusion of structural and energetic signals into interpretable maps of sequestration risk, off-target exposure, and intracellular sink formation.
By replacing narrow safety panels with a global interaction map, AgnostiMap exposes hidden binding reservoirs that drive hepatic accumulation and unpredictable clinical toxicity ← the true blind spots of modern drug discovery.
Platform Specifications
Proteome Coverage: 20,000+ human proteins (AlphaFold v3–derived structures)
Ligand Scope: < 800 Da (small-molecule optimized)
Dynamic Validation: Molecular dynamics simulations using OpenMM
End-to-End Docking Workflow